Decoding the origin of life with lost biochemical clues
Metabolism is the “beating heart of the cell”. New research from ELSI traces the history of metabolism from the primordial Earth to modern times (left to right). The history of compound discovery over time (white line) is cyclical, almost EKG-like. Credit: NASA’s Goddard Space Flight Center/Francis Reddy/NASA/ESA
A new study shows that only a few “forgotten” biochemical reactions are needed to transform simple geochemical compounds into the complex molecules of life.
The origin of life on Earth has long been a mystery that eluded scientists. The key question is how much of the history of life on Earth has been lost to time. It’s quite common for a single person species be “phased out” by a biochemical reaction, and if this happens in enough species, such reactions could be “forgotten” by life on Earth. But if the history of biochemistry is littered with forgotten reactions, would there be a way to tell?
This question inspired researchers from the Earth-Life Science Institute (ELSI) at the Tokyo Institute of Technology and the California Institute of Technology (CalTech) in the US. They believed that the forgotten chemistry would appear as discontinuities or “breaks” in the path taken by chemistry from simple geochemical molecules to complex biological molecules.
The evolution of Earth’s early biochemistry
The early Earth was rich in simple compounds such as hydrogen sulfide, ammonia, and carbon dioxide—molecules not usually associated with sustaining life. But, billions of years ago, early life relied on these simple molecules as a source of raw materials. As life evolved, biochemical processes gradually transformed these precursors into the compounds that are still found today. These processes represent the earliest metabolic pathways.
To construct a model of the evolutionary history of metabolism at the biosphere level, the research team compiled a database of 12,262 biochemical reactions from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Credit: Goldford, JE, Nat Ecol Evol (2024)
Biochemical evolution research methodology
To model the history of biochemistry, the ELSI researchers—Specially Appointed Associate Professor Harrison B. Smith, Specially Appointed Associate Professor Liam M. Longo, and Associate Professor Shawn Erin McGlynn, in collaboration with research scientist Joshua Goldford of CalTech—needed an inventory of all known biochemical reactions, in order to understand what kinds of chemistry life can perform.
They turned to the Kyoto Encyclopedia of Genes and Genomes database, which cataloged more than 12,000 biochemical reactions. With the reactions in hand, they began to model the gradual development of metabolism.
Challenges in modeling metabolic evolution
Previous attempts to model the evolution of metabolism in this way have consistently failed to produce the most widespread, complex molecules used by modern life. However, the reason was not entirely clear. As before, when the researchers ran their model, they found that only a few compounds could be produced.
One way to get around this problem is to boost stagnant chemistry by hand-injecting modern compounds. The researchers decided on a different approach: they wanted to determine how much reactions were missing. And their search led them back to one of the most important molecules in all of biochemistry: adenosine triphosphate (ATP).
The ATP bottleneck and its resolution
ATP is the cell’s energy currency because it can be used to drive reactions—like building proteins—that wouldn’t otherwise occur in water. ATP, however, has a unique property: Reactions that generate ATP themselves require ATP. In other words, unless ATP is already present, there is no other way for life today to create ATP. This cyclical dependence was the reason why the model stopped.
How could this “ATP bottleneck” be solved? As it turns out, the reactive part of ATP is remarkably similar to the inorganic compound polyphosphate. By allowing the ATP-generating reactions to use polyphosphate instead of ATP—by modifying only eight reactions in all—almost all of modern basic metabolism could be achieved. The researchers were then able to estimate the relative ages of all common metabolites and ask sharp questions about the history of metabolic pathways.
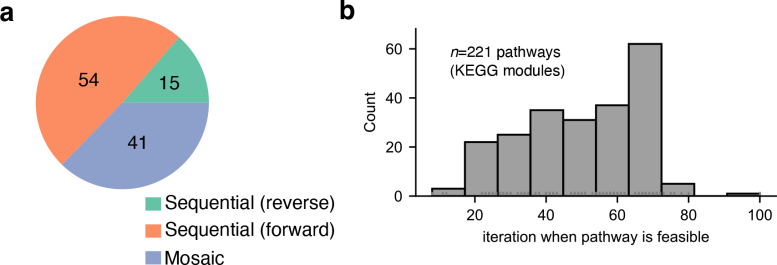
Metabolic pathways: linear versus mosaic
One such question is whether biological pathways were built in a linear fashion—in which one reaction after another is added in a sequential fashion—or whether the pathway reactions emerged as a mosaic, in which reactions of very different ages were joined together to form something new. The researchers were able to quantify this, finding that both types of pathways are almost equally common throughout metabolism.
Conclusion and implications
But back to the question that inspired the study – how much biochemistry has been lost to time? “We may never know for sure, but our research has provided an important piece of evidence: only eight new reactions, all of which resemble common biochemical reactions, are needed to bridge geochemistry and biochemistry,” says Smith.
“This does not prove that the space of missing biochemistry is small, but it does show that even reactions that have become extinct can be rediscovered from traces left in modern biochemistry,” concludes Smith.
Reference: “Primitive Purine Biosynthesis Links Ancient Geochemistry to Modern Metabolism” Joshua E. Goldford, Harrison B. Smith, Liam M. Longo, Boswell A. Wing, and Shawn Erin McGlynn, 22 March 2024, Ecology of nature and evolution.
DOI: 10.1038/s41559-024-02361-4
Related Posts

Kilauea, the active Hawaiian volcano, could erupt like a ‘toy rocket’, a new study suggests

Surprising evolutionary insights revealed by first complete great ape chromosome sequences
